Make separate plots for each model compartment. Assumes model output is structured
as that produced from solve_ode.
plot_model(sim, prev_sim = NULL, model_labels = NULL, facet = TRUE)Arguments
- sim
A tibble of model output as formatted by
solve_ode. Optionally a list of simulations can be passed when comparing multiple model runs.- prev_sim
A second tibble of model output formatted as for
sim. Used to compare to model runs. Can only be supplied ifsimis not a list.- model_labels
A character vector of model names. Defaults to
c("Current", "Previous")when two model simulations are used and the list names whensimis a list. Ifsimis unnamed the index of the list is used.- facet
Logical, defaults to
TRUE. IfFALSEthen the plot will not be faceted otherwise it will be.
Value
A Plot of each model compartments population over time.
Examples
## Intialise
N = 100000
I_0 = 1
S_0 = N - I_0
R_0 = 1.1
beta = R_0
##Time for model to run over
tbegin = 0
tend = 50
times <- seq(tbegin, tend, 1)
##Vectorise input
parameters <- as.matrix(c(beta = beta))
inits <- as.matrix(c(S = S_0, I = I_0))
sim <- solve_ode(model = SI_ode, inits, parameters, times, as.data.frame = TRUE)
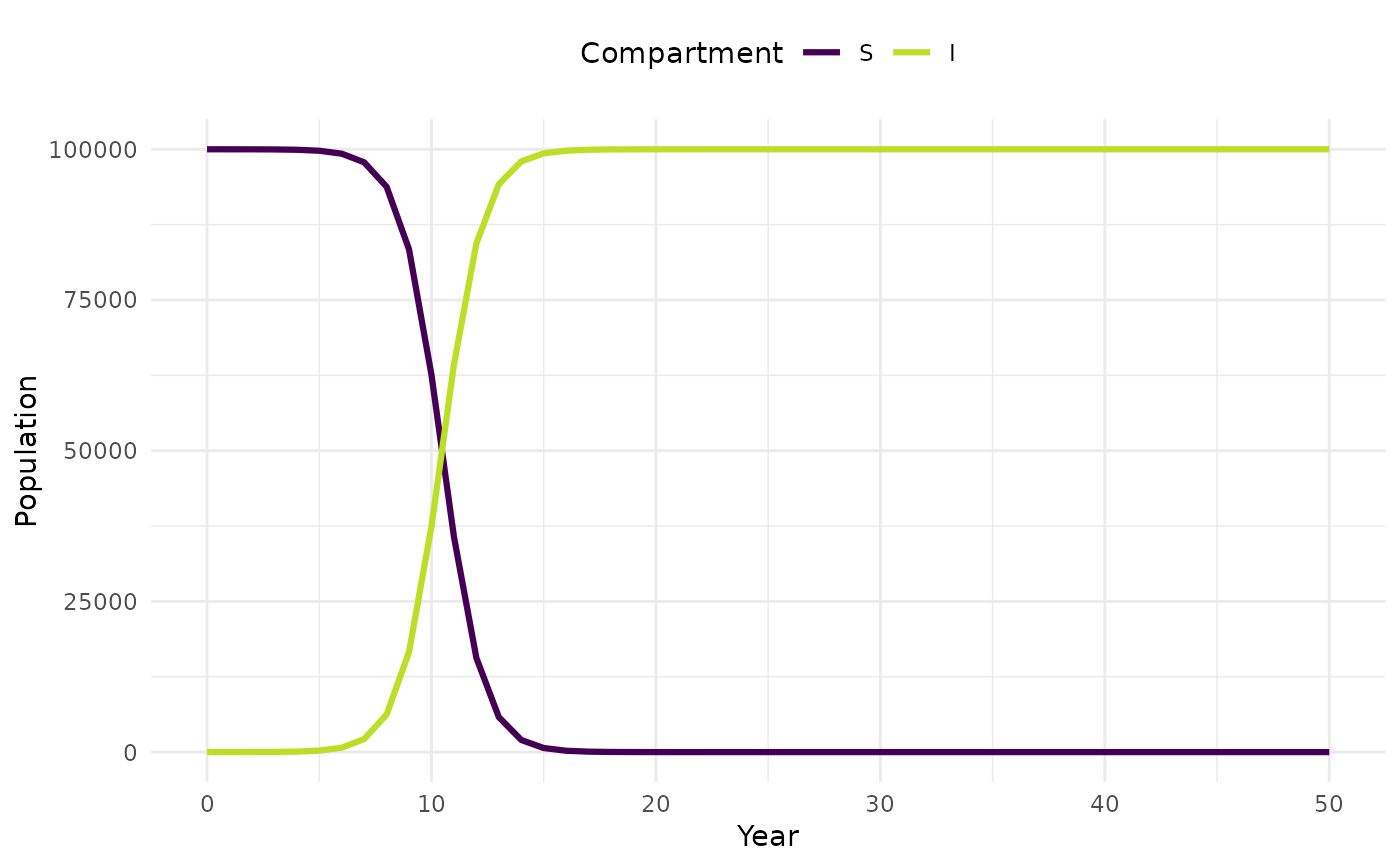
plot_model(sim, facet = FALSE)
#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.
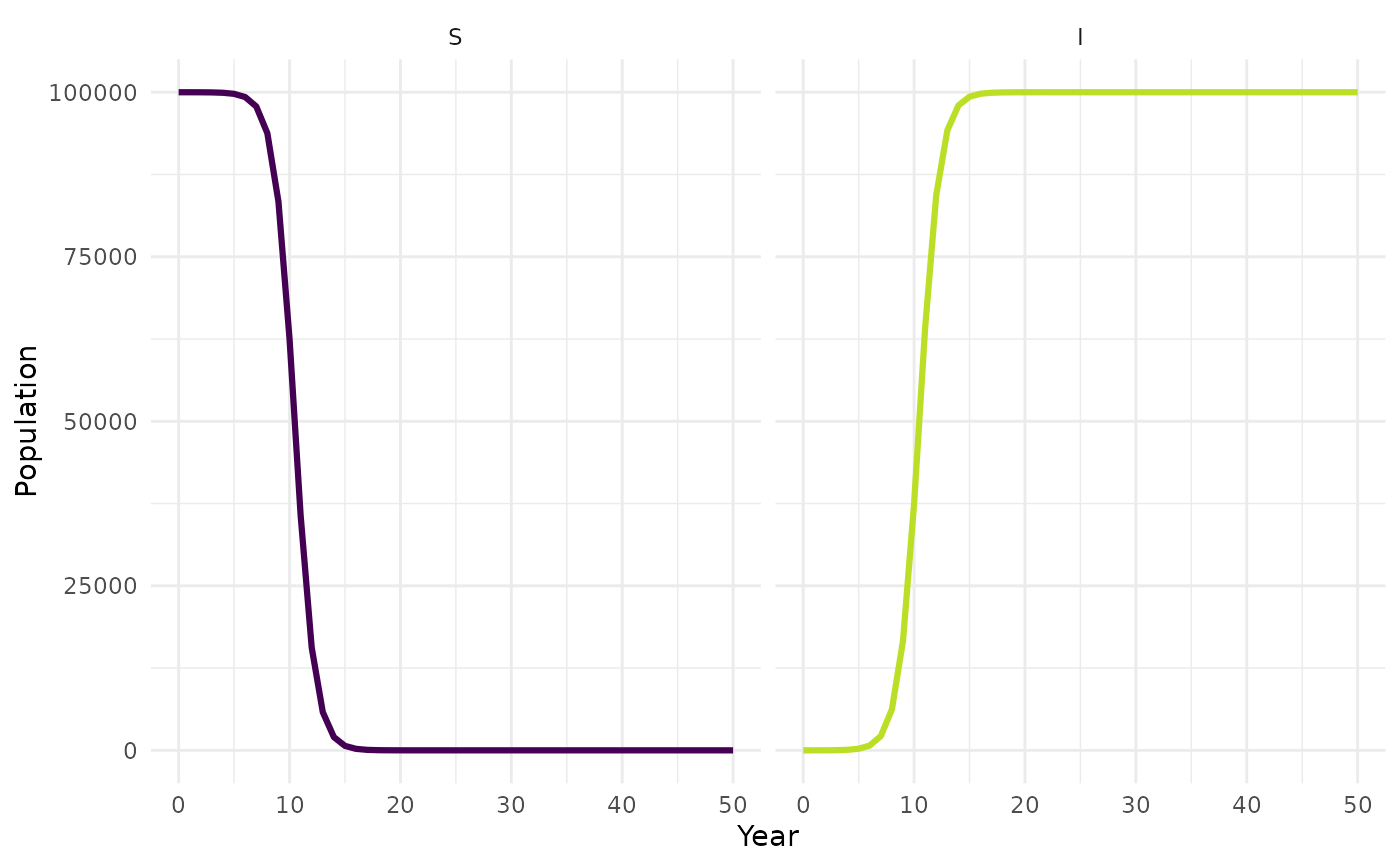
 plot_model(sim, facet = TRUE)
#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.
#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.
plot_model(sim, facet = TRUE)
#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.
#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.
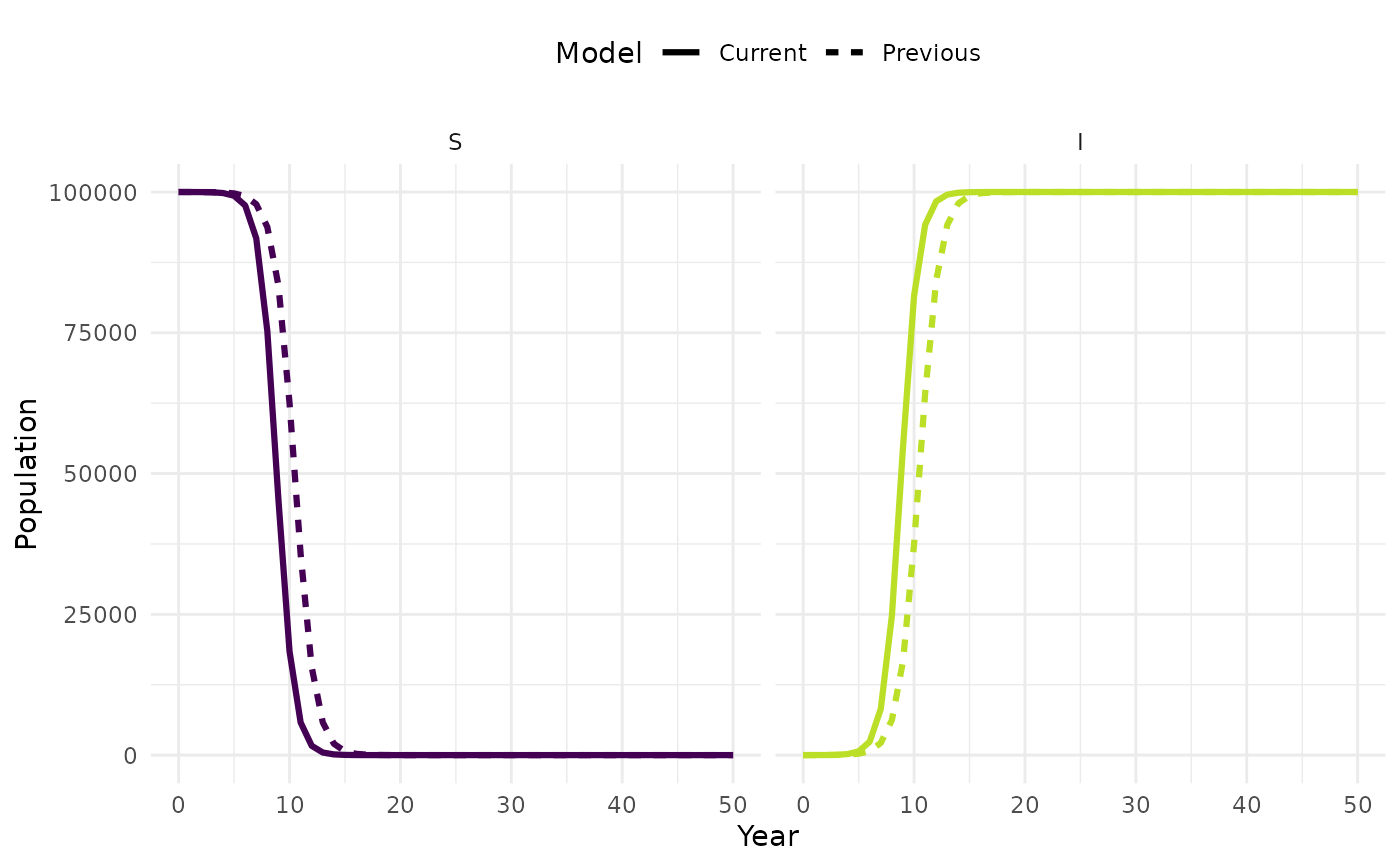
 ## Compare with an updated model run
#'## Intialise
R_0 = 1.3
beta = R_0
parameters <- as.matrix(c(beta = beta))
new_sim <- solve_ode(model = SI_ode, inits, parameters, times, as.data.frame = TRUE)
plot_model(new_sim,sim, facet = FALSE)
## Compare with an updated model run
#'## Intialise
R_0 = 1.3
beta = R_0
parameters <- as.matrix(c(beta = beta))
new_sim <- solve_ode(model = SI_ode, inits, parameters, times, as.data.frame = TRUE)
plot_model(new_sim,sim, facet = FALSE)
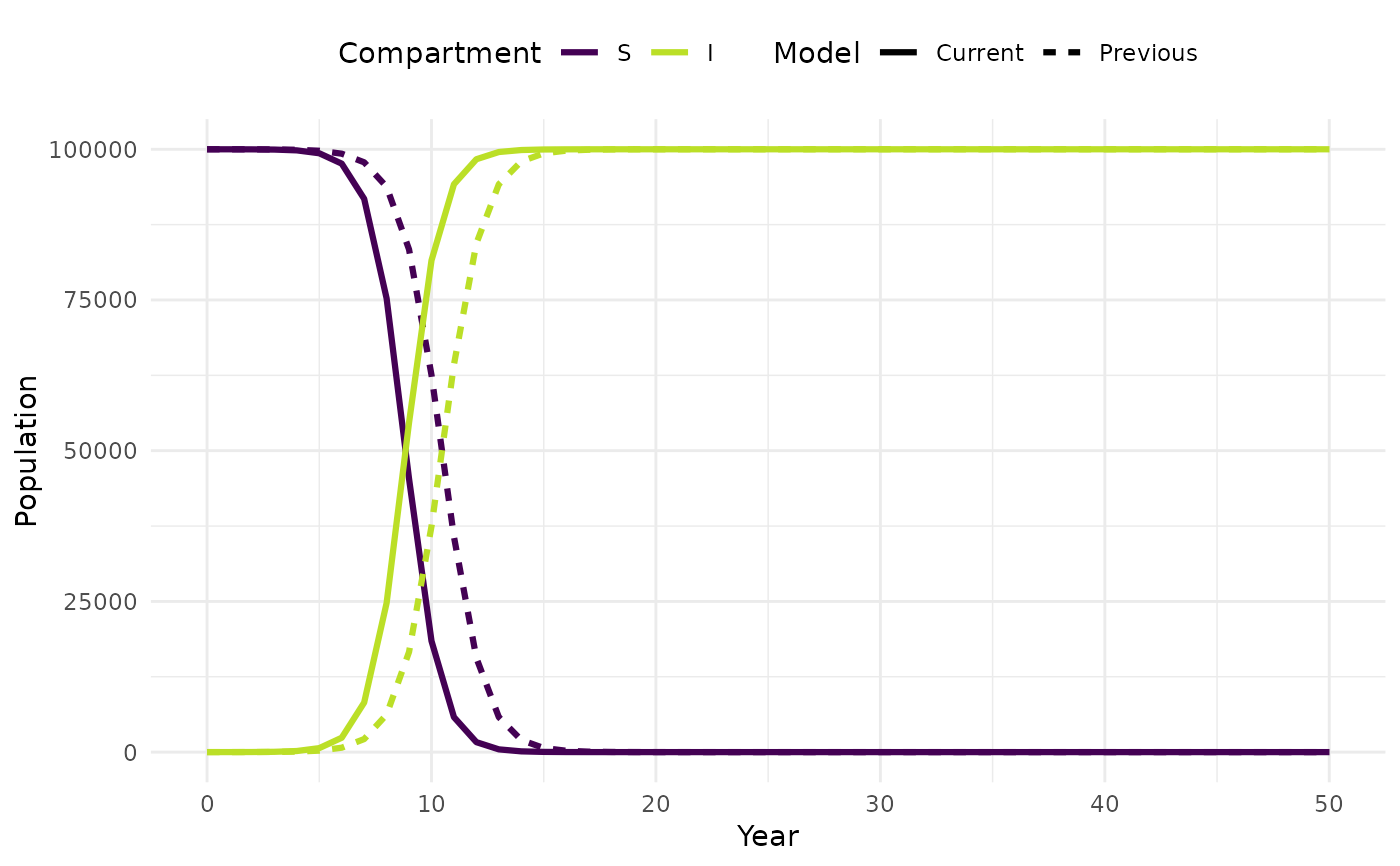 plot_model(new_sim, sim, facet = TRUE)
#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.
plot_model(new_sim, sim, facet = TRUE)
#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.
 ## Passing in the simulations as a list
plot_model(list("Current" = new_sim, "Previous" = sim), facet = TRUE)
#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.
## Passing in the simulations as a list
plot_model(list("Current" = new_sim, "Previous" = sim), facet = TRUE)
#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.